
samtools view -hL regions.bed aln.bam > aln_filtered. Samtools does have an option to filter the reads according to regions specified in the BED format but it will not automatically annotate them. You can use any scripting language like awk or perl to parse. See this link for the details on the SAM format. What I would do is convert the BAM to SAM (using samtools) or to BED (using bedtools: bamtobed) and parse the columns of the SAM/ BED file describing the read locations with the GTF file.
#SEQUENCHER COMPANY INSTALL#
I don't have much experience with this tool. Create a shortcut of 'C:/Program Files(x86)/Gene Codes/Sequencher 4.7/Sequencher.exe' and place the shortcut on the desktop Celebrate a complete installation of Sequencher MacOS: Download dmg installer Install Mock your windows using co-workers about how easy to install Sequencher was. Sequencher has integrated the comprehensive Cufflinks suite for in-depth transcript analysis and differential gene expression of your RNA-Seq data. (SPF) Find, read and cite all the research you need on ResearchGate.
#SEQUENCHER COMPANY PDF#
You can obtain the association between genomic locations and the reads using the BEDOPS toolkit as mentioned in this Biostars post. PDF Sequence analysis of integrated-telomere (166 bp) controls in rad52 for S2 Fig. The annotations are available in the form of GTF/GFF files. This information is not available unless you have the genome annotations. Free Download and information on Sequencher - Sequencher is the industry standard software for DNA sequence analysis. You want to know, to which genes do these reads map to. Samtools is a command-line program that can help you extract information from the aln.bam file. As far as I know, sequencher is used to clean and assemble Sanger sequencing data, but my experience with that program is almost 10 years old.

Since you already have the alignments ( aln.bam) to hg19, you do not need to perform alignment. IGV will also use the file to help it find the aligned reads in the aln.bam file. Again, there are many aligners available Bowtie, STAR and now a new one called HISAT. Then you have to align your trimmed reads to a reference, using an aligner. You can find the adaptor composition of each read by subtracting the read length post trimming from the read length pre-trimming (this would be same for all reads). You can get to know the adaptor sequence by running FastQC. FastQC is for analysing the read qualities and average composition.
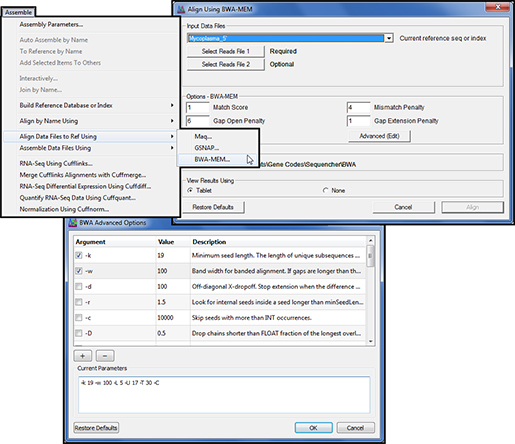
Samtools is not an aligner it is used to analyse alignments.


 0 kommentar(er)
0 kommentar(er)
